Open Sentinel-3-OLCI-Level-1-EFR Data with GDAL and eopfzarr

This Jupyter notebook demonstrates how to open Sentinel-3-OLCI-Level-1-EFR data using GDAL, the eopfzarr plugin, mirroring the structure and functionality of the xarray-eopf example notebook for Sentinel-3.
Key features demonstrated:
GDAL native approach: Direct access to Zarr subdatasets
xarray-style visualization: Simple plotting with
robust=TrueGeographic coordinates: Latitude/longitude access like xarray-eopf analysis mode
Multi-product exploration: OLCI L1/L2 and SLSTR products
Products covered:
Sentinel-3 OLCI Level-1 EFR (TOA radiances, full resolution)
Table of Contents¶
Run this notebook interactively with all dependencies pre-installed
Setup and Imports¶
Import the necessary libraries for working with GDAL, xarray, and visualization tools.
from osgeo import gdal
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
# Enable GDAL exceptions
gdal.UseExceptions()
# Helper function for eopfzarr paths (remote access)
def eopfzarr_path(url, subdataset=None):
base = f'EOPFZARR:"/vsicurl/{url}"'
return f"{base}:{subdataset}" if subdataset else base
# Sample URLs for Sentinel-3 products
olci_l1_efr_url = "https://objects.eodc.eu/e05ab01a9d56408d82ac32d69a5aae2a:202508-s03olcefr/19/products/cpm_v256/S3B_OL_1_EFR____20250819T074058_20250819T074358_20250819T092155_0179_110_106_3420_ESA_O_NR_004.zarr"Explore OLCI L1 EFR Structure¶
Open the Sentinel-3 OLCI Level-1 EFR product and explore its structure, including available subdatasets.
# Open L1 EFR root to explore structure
l1_efr_root_ds = gdal.Open(eopfzarr_path(olci_l1_efr_url))
print(f"Driver: {l1_efr_root_ds.GetDriver().ShortName}")
print(f"Projection (CRS): {l1_efr_root_ds.GetProjection()}")
print(f"GeoTransform: {l1_efr_root_ds.GetGeoTransform()}")
# List subdatasets (hierarchical groups like /measurements/oa01_radiance)
subdatasets = l1_efr_root_ds.GetMetadata("SUBDATASETS")
print("Available Subdatasets for OLCI L1 EFR:")
for key, value in subdatasets.items():
if key.startswith("SUBDATASET_") and "_DESC" not in key:
print(f" {value.split(':')[-1]}")
l1_efr_root_ds = NoneDriver: EOPFZARR
Projection (CRS): GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AXIS["Latitude",NORTH],AXIS["Longitude",EAST],AUTHORITY["EPSG","4326"]]
GeoTransform: (18.4535, 0.031338476563, 0.0, -21.0585, 0.0, -0.02630859375)
Available Subdatasets for OLCI L1 EFR:
/conditions/geometry/latitude
/conditions/geometry/longitude
/conditions/geometry/oaa
/conditions/geometry/oza
/conditions/geometry/saa
/conditions/geometry/sza
/conditions/image/altitude
/conditions/image/detector_index
/conditions/image/frame_offset
/conditions/image/latitude
/conditions/image/longitude
/conditions/instrument/fwhm
/conditions/instrument/lambda0
/conditions/instrument/relative_spectral_covariance
/conditions/instrument/solar_flux
/conditions/meteorology/atmospheric_temperature_profile
/conditions/meteorology/horizontal_wind
/conditions/meteorology/humidity
/conditions/meteorology/sea_level_pressure
/conditions/meteorology/total_columnar_water_vapour
/conditions/meteorology/total_ozone
/conditions/orphans/altitude
/conditions/orphans/detector_index
/conditions/orphans/frame_offset
/conditions/orphans/latitude
/conditions/orphans/longitude
/conditions/orphans/sza
/measurements/latitude
/measurements/longitude
/measurements/oa01_radiance
/measurements/oa02_radiance
/measurements/oa03_radiance
/measurements/oa04_radiance
/measurements/oa05_radiance
/measurements/oa06_radiance
/measurements/oa07_radiance
/measurements/oa08_radiance
/measurements/oa09_radiance
/measurements/oa10_radiance
/measurements/oa11_radiance
/measurements/oa12_radiance
/measurements/oa13_radiance
/measurements/oa14_radiance
/measurements/oa15_radiance
/measurements/oa16_radiance
/measurements/oa17_radiance
/measurements/oa18_radiance
/measurements/oa19_radiance
/measurements/oa20_radiance
/measurements/oa21_radiance
/measurements/orphans/altitude
/measurements/orphans/latitude
/measurements/orphans/longitude
/measurements/orphans/oa01_radiance
/measurements/orphans/oa02_radiance
/measurements/orphans/oa03_radiance
/measurements/orphans/oa04_radiance
/measurements/orphans/oa05_radiance
/measurements/orphans/oa06_radiance
/measurements/orphans/oa07_radiance
/measurements/orphans/oa08_radiance
/measurements/orphans/oa09_radiance
/measurements/orphans/oa10_radiance
/measurements/orphans/oa11_radiance
/measurements/orphans/oa12_radiance
/measurements/orphans/oa13_radiance
/measurements/orphans/oa14_radiance
/measurements/orphans/oa15_radiance
/measurements/orphans/oa16_radiance
/measurements/orphans/oa17_radiance
/measurements/orphans/oa18_radiance
/measurements/orphans/oa19_radiance
/measurements/orphans/oa20_radiance
/measurements/orphans/oa21_radiance
/quality/latitude
/quality/longitude
/quality/oa01_radiance_unc
/quality/oa02_radiance_unc
/quality/oa03_radiance_unc
/quality/oa04_radiance_unc
/quality/oa05_radiance_unc
/quality/oa06_radiance_unc
/quality/oa07_radiance_unc
/quality/oa08_radiance_unc
/quality/oa09_radiance_unc
/quality/oa10_radiance_unc
/quality/oa11_radiance_unc
/quality/oa12_radiance_unc
/quality/oa13_radiance_unc
/quality/oa14_radiance_unc
/quality/oa15_radiance_unc
/quality/oa16_radiance_unc
/quality/oa17_radiance_unc
/quality/oa18_radiance_unc
/quality/oa19_radiance_unc
/quality/oa20_radiance_unc
/quality/oa21_radiance_unc
/quality/quality_flags
/quality/orphans/altitude
/quality/orphans/latitude
/quality/orphans/longitude
/quality/orphans/oa01_radiance_unc
/quality/orphans/oa02_radiance_unc
/quality/orphans/oa03_radiance_unc
/quality/orphans/oa04_radiance_unc
/quality/orphans/oa05_radiance_unc
/quality/orphans/oa06_radiance_unc
/quality/orphans/oa07_radiance_unc
/quality/orphans/oa08_radiance_unc
/quality/orphans/oa09_radiance_unc
/quality/orphans/oa10_radiance_unc
/quality/orphans/oa11_radiance_unc
/quality/orphans/oa12_radiance_unc
/quality/orphans/oa13_radiance_unc
/quality/orphans/oa14_radiance_unc
/quality/orphans/oa15_radiance_unc
/quality/orphans/oa16_radiance_unc
/quality/orphans/oa17_radiance_unc
/quality/orphans/oa18_radiance_unc
/quality/orphans/oa19_radiance_unc
/quality/orphans/oa20_radiance_unc
/quality/orphans/oa21_radiance_unc
/quality/orphans/quality_flags
Simple Plotting with xarray-style¶
Create simple plots using xarray-style syntax, similar to the xarray-eopf approach.
def gdal_to_xarray(url, subdataset, size=4000):
"""Convert GDAL data to xarray DataArray for simple plotting like xarray-eopf"""
path = eopfzarr_path(url, subdataset)
ds = gdal.Open(path)
band = ds.GetRasterBand(1)
data = band.ReadAsArray(0, 0, size, size).astype(np.float32)
nodata = band.GetNoDataValue()
# Create simple coordinate arrays
y_coords = np.arange(size)
x_coords = np.arange(size)
# Create xarray DataArray
da = xr.DataArray(
data,
coords={"y": y_coords, "x": x_coords},
dims=["y", "x"],
name=subdataset.split("/")[-1],
)
# Handle NoData
if nodata is not None:
da = da.where(da != nodata)
ds = None
return da
# Simple plotting like xarray-eopf: ds.oa01_radiance[::4, ::4].plot(robust=True)
print("Creating xarray-style plots similar to xarray-eopf notebook...")
# One-liner equivalent to xarray-eopf
gdal_to_xarray(olci_l1_efr_url, "measurements/oa01_radiance")[::4, ::4].plot(
robust=True, figsize=(8, 6)
)
plt.title(
"OLCI L1 EFR Oa01 Radiance\n(xarray-style: robust=True, subsampled [::4, ::4])"
)
plt.show()
print(
"✅ This is equivalent to: ds.oa01_radiance[::4, ::4].plot(robust=True) in xarray-eopf"
)Creating xarray-style plots similar to xarray-eopf notebook...
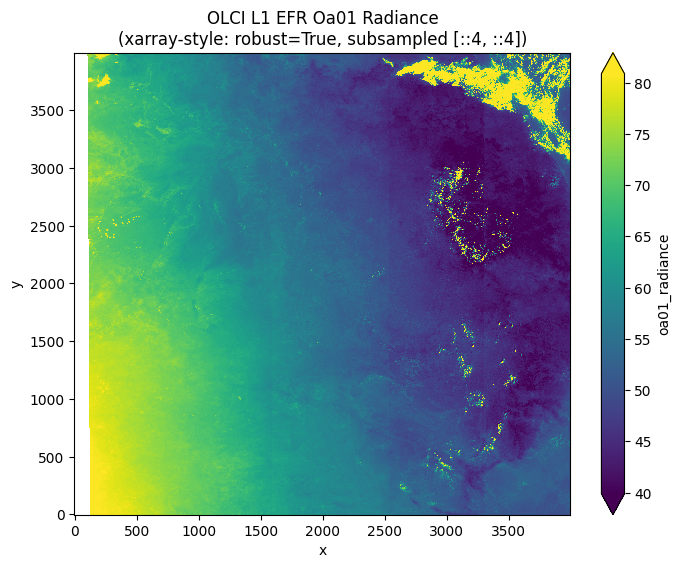
✅ This is equivalent to: ds.oa01_radiance[::4, ::4].plot(robust=True) in xarray-eopf
Geographic Coordinates: Latitude and Longitude¶
Sentinel-3 OLCI products include latitude and longitude coordinates for each pixel. Let’s explore how to access and visualize these with GDAL eopfzarr, similar to the xarray-eopf analysis mode.
# First, let's explore what coordinate datasets are available
print("Exploring available coordinate datasets in OLCI L1 EFR...")
# Open the root to see what coordinate groups are available
l1_efr_root_ds = gdal.Open(eopfzarr_path(olci_l1_efr_url))
subdatasets = l1_efr_root_ds.GetMetadata("SUBDATASETS")
# Look for coordinate-related subdatasets
coord_datasets = []
for key, value in subdatasets.items():
if key.startswith("SUBDATASET_") and "_DESC" not in key:
dataset_path = value.split(":")[-1]
if any(
coord in dataset_path.lower()
for coord in ["lat", "lon", "coordinate", "geometry"]
):
coord_datasets.append(dataset_path)
print("Available coordinate datasets:")
for coord in coord_datasets:
print(f" {coord}")
l1_efr_root_ds = NoneExploring available coordinate datasets in OLCI L1 EFR...
Available coordinate datasets:
/conditions/geometry/latitude
/conditions/geometry/longitude
/conditions/geometry/oaa
/conditions/geometry/oza
/conditions/geometry/saa
/conditions/geometry/sza
/conditions/image/latitude
/conditions/image/longitude
/conditions/instrument/relative_spectral_covariance
/conditions/orphans/latitude
/conditions/orphans/longitude
/measurements/latitude
/measurements/longitude
/measurements/orphans/latitude
/measurements/orphans/longitude
/quality/latitude
/quality/longitude
/quality/orphans/latitude
/quality/orphans/longitude
def gdal_to_xarray_with_coords(
url, data_subdataset, lat_subdataset, lon_subdataset, size=1000
):
"""Convert GDAL data to xarray DataArray with geographic coordinates"""
# Load the data
data_path = eopfzarr_path(url, data_subdataset)
data_ds = gdal.Open(data_path)
data_band = data_ds.GetRasterBand(1)
data = data_band.ReadAsArray(0, 0, size, size).astype(np.float32)
data_nodata = data_band.GetNoDataValue()
data_ds = None
# Load latitude coordinates
lat_path = eopfzarr_path(url, lat_subdataset)
lat_ds = gdal.Open(lat_path)
lat_band = lat_ds.GetRasterBand(1)
lat = lat_band.ReadAsArray(0, 0, size, size).astype(np.float32)
lat_ds = None
# Load longitude coordinates
lon_path = eopfzarr_path(url, lon_subdataset)
lon_ds = gdal.Open(lon_path)
lon_band = lon_ds.GetRasterBand(1)
lon = lon_band.ReadAsArray(0, 0, size, size).astype(np.float32)
lon_ds = None
# Create xarray DataArray with geographic coordinates
da = xr.DataArray(
data,
coords={
"y": np.arange(size),
"x": np.arange(size),
"latitude": (("y", "x"), lat),
"longitude": (("y", "x"), lon),
},
dims=["y", "x"],
name=data_subdataset.split("/")[-1],
)
# Handle NoData
if data_nodata is not None:
da = da.where(da != data_nodata)
return da
# Load data with geographic coordinates using EOPF measurements paths
print("Loading Oa01 radiance with geographic coordinates...")
print("Using measurements group paths: /measurements/latitude, /measurements/longitude")
try:
oa01_with_coords = gdal_to_xarray_with_coords(
olci_l1_efr_url,
"measurements/oa01_radiance",
"measurements/latitude",
"measurements/longitude",
size=800, # Moderate size for demo
)
# Display coordinate information
print("\n✓ Successfully loaded geographic coordinates!")
print(f" Data shape: {oa01_with_coords.shape}")
print(
f" Latitude range: {float(oa01_with_coords.latitude.min()):.3f} to {float(oa01_with_coords.latitude.max()):.3f}"
)
print(
f" Longitude range: {float(oa01_with_coords.longitude.min()):.3f} to {float(oa01_with_coords.longitude.max()):.3f}"
)
coord_success = True
except Exception as e:
print(f"Primary coordinates failed: {e}")
# Try orphans coordinates as fallback
try:
print(
"Trying fallback: /measurements/orphans/latitude, /measurements/orphans/longitude"
)
oa01_with_coords = gdal_to_xarray_with_coords(
olci_l1_efr_url,
"measurements/oa01_radiance",
"measurements/orphans/latitude",
"measurements/orphans/longitude",
size=800,
)
print("✓ Successfully loaded orphans coordinates!")
coord_success = True
except Exception as e2:
print(f"Orphans coordinates also failed: {e2}")
coord_success = FalseLoading Oa01 radiance with geographic coordinates...
Using measurements group paths: /measurements/latitude, /measurements/longitude
✓ Successfully loaded geographic coordinates!
Data shape: (800, 800)
Latitude range: -23.643 to -21.059
Longitude range: 21.465 to 24.124
# Geographic visualization - equivalent to xarray-eopf analysis mode
if coord_success:
print("Demonstrating geographic plotting - xarray-eopf analysis mode equivalent:")
print(
"xarray-eopf: ds.oa01_radiance[::4, ::4].plot(x='longitude', y='latitude', robust=True)"
)
print("Our GDAL eopfzarr equivalent:")
# Plot the data with geographic coordinates
fig, axes = plt.subplots(2, 2, figsize=(16, 12))
# 1. Data with array indices (standard)
oa01_with_coords[::4, ::4].plot(ax=axes[0, 0], robust=True, cmap="viridis")
axes[0, 0].set_title("Oa01 Radiance\n(Array indices)")
# 2. Latitude field
oa01_with_coords.latitude[::4, ::4].plot(ax=axes[0, 1], cmap="coolwarm")
axes[0, 1].set_title("Latitude Coordinates\n(degrees North)")
# 3. Longitude field
oa01_with_coords.longitude[::4, ::4].plot(ax=axes[1, 0], cmap="coolwarm")
axes[1, 0].set_title("Longitude Coordinates\n(degrees East)")
# 4. Geographic plot with lat/lon as axes (like xarray-eopf analysis mode)
try:
subset = oa01_with_coords[::6, ::6] # Subsample for clarity
lat_2d = subset.latitude.values
lon_2d = subset.longitude.values
data_2d = subset.values
im = axes[1, 1].pcolormesh(
lon_2d, lat_2d, data_2d, cmap="viridis", shading="auto"
)
axes[1, 1].set_xlabel("Longitude (degrees East)")
axes[1, 1].set_ylabel("Latitude (degrees North)")
axes[1, 1].set_title("Oa01 Radiance\n(Geographic coordinates)")
plt.colorbar(im, ax=axes[1, 1])
except Exception as e:
axes[1, 1].text(
0.5,
0.5,
f"Geographic plot failed:\n{str(e)[:50]}...",
ha="center",
va="center",
transform=axes[1, 1].transAxes,
)
axes[1, 1].set_title("Geographic plot (attempted)")
plt.tight_layout()
plt.show()
print(
"✅ Successfully demonstrated geographic coordinate access with GDAL eopfzarr!"
)
print(
"✅ Coordinates directly accessible from /measurements/latitude and /measurements/longitude"
)
else:
print("Geographic coordinate loading failed. Check available coordinate datasets.")Demonstrating geographic plotting - xarray-eopf analysis mode equivalent:
xarray-eopf: ds.oa01_radiance[::4, ::4].plot(x='longitude', y='latitude', robust=True)
Our GDAL eopfzarr equivalent:
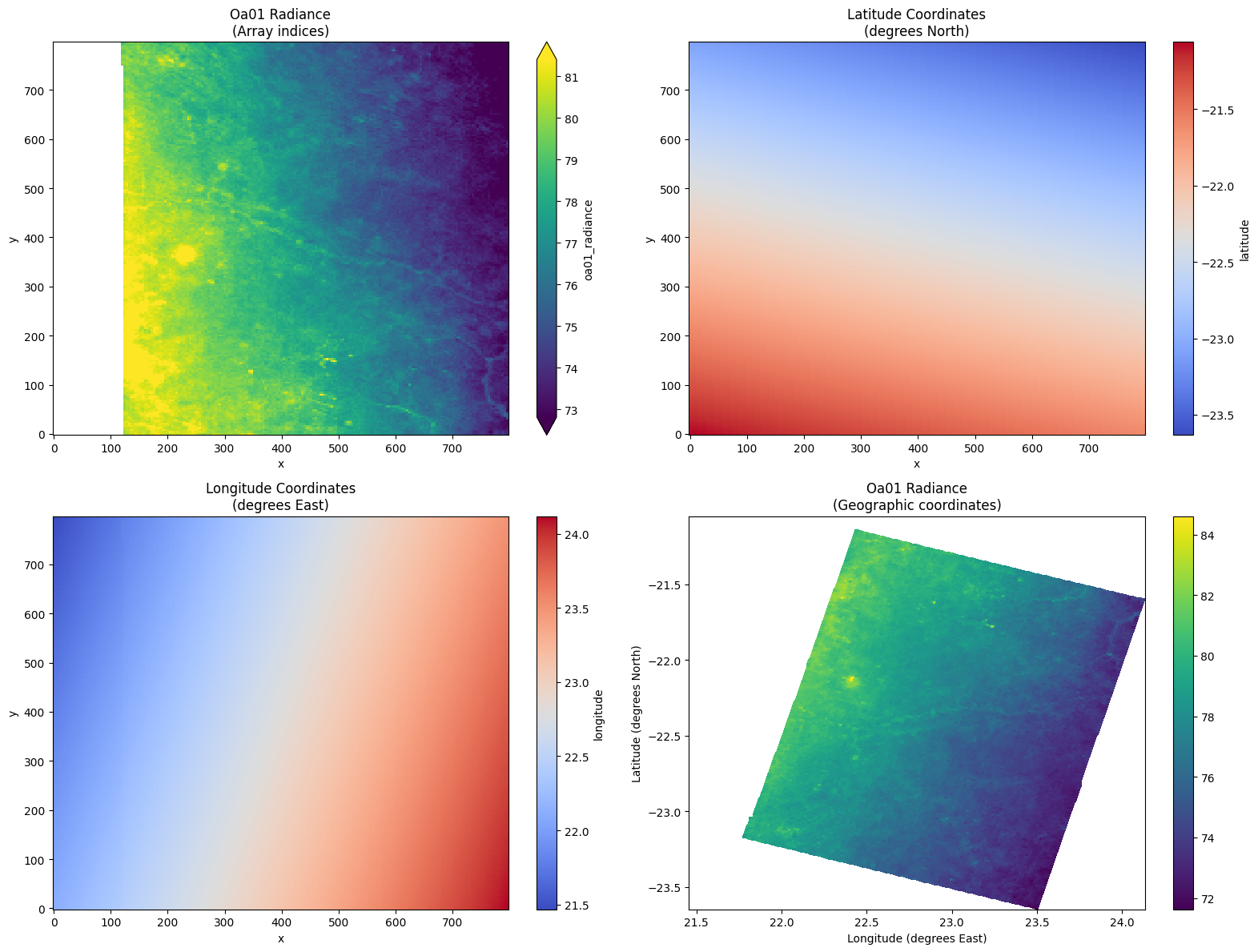
✅ Successfully demonstrated geographic coordinate access with GDAL eopfzarr!
✅ Coordinates directly accessible from /measurements/latitude and /measurements/longitude
# Simple geographic plotting - one-liner equivalent to xarray-eopf
if coord_success:
print("\nSimple geographic plotting (xarray-eopf style one-liner equivalent):")
# Create a simple geographic plot
plt.figure(figsize=(14, 10))
# Use the geographic coordinates for plotting
subset = oa01_with_coords[::8, ::8] # Subsample for performance
lat_vals = subset.latitude.values
lon_vals = subset.longitude.values
data_vals = subset.values
# Geographic plot - equivalent to ds.oa01_radiance.plot(x='longitude', y='latitude', robust=True)
plt.pcolormesh(lon_vals, lat_vals, data_vals, cmap="viridis", shading="auto")
plt.colorbar(label="Oa01 Radiance (mW/m²/sr/nm)")
plt.xlabel("Longitude (degrees East)")
plt.ylabel("Latitude (degrees North)")
plt.title(
"OLCI L1 EFR Oa01 Radiance\n(Geographic coordinates - xarray-eopf analysis mode equivalent)"
)
plt.grid(True, alpha=0.3)
plt.show()
print(
"✅ GDAL eopfzarr successfully replicates xarray-eopf analysis mode geographic plotting!"
)
print(
"✅ Direct access to lat/lon from measurements group enables full geographic analysis"
)
else:
print("Geographic plotting requires successful coordinate loading.")
Simple geographic plotting (xarray-eopf style one-liner equivalent):
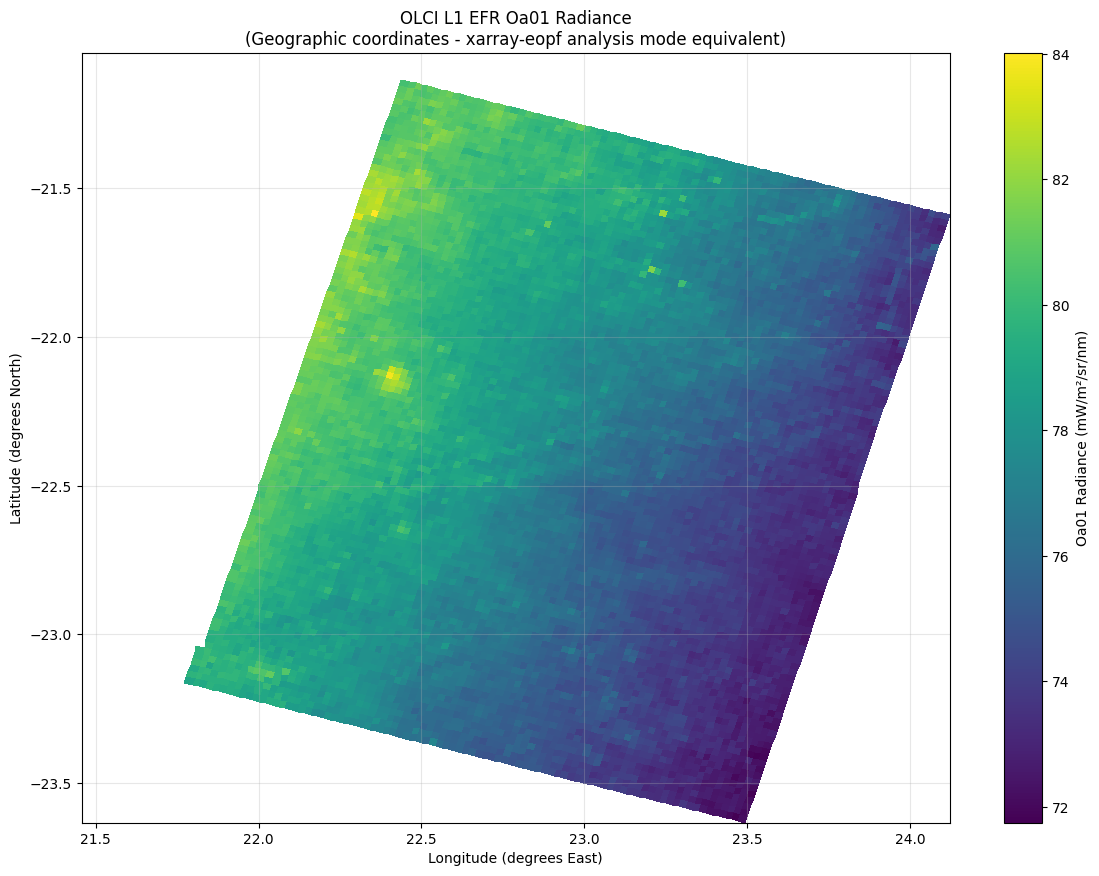
✅ GDAL eopfzarr successfully replicates xarray-eopf analysis mode geographic plotting!
✅ Direct access to lat/lon from measurements group enables full geographic analysis
Geographic Coordinate Comparison: GDAL vs xarray-eopf¶
xarray-eopf analysis mode approach:
# Analysis mode automatically handles coordinates
ds = xr.open_dataset(url, engine="eopf-zarr", op_mode="analysis")
ds.oa01_radiance.plot(x='longitude', y='latitude', robust=True)
# Manual coordinate loading and geographic plotting
da = gdal_to_xarray_with_coords(olci_l1_efr_url, 'measurements/oa01_radiance', 'measurements/latitude', 'measurements/longitude')
plt.pcolormesh(da.longitude, da.latitude, da.values)