Random Forest Based Land-Cover Mapping with Sentinel-2
Demonstrate how to perform a basic land cover mapping using random forest and the Sentinel-2 data in EOPF Zarr format

Table of Contents¶
Run this notebook interactively with all dependencies pre-installed
Introduction¶
In this notebook, we will explore land cover mapping, a practice that has been integral to remote sensing since its inception. Land cover products are essential for studying the functional and morphological changes in Earth’s ecosystems and the environment, playing a crucial role in understanding climate change and carbon circulation (Congalton et al., 2014; Feddema et al., 2005; Sellers et al., 1997). Additionally, land cover mapping provides valuable information for policy development and a wide range of applications within the natural and life sciences, making it one of the most extensively studied applications in remote sensing (Yu et al., 2014; Tucker et al., 1985; Running, 2008; Yang et al., 2013). The diverse applications of land cover mapping lead to varied user needs. Depending on the specific use case, there can be significant differences in the desired target labels, the requested target year(s), the required output resolution, the feature set used, the stratification strategy employed, and more.
This exercise demonstrates how to efficiently use the Sentinel-2 EOPF Zarr products to implement a basic land cover classification.
Install the missing libraries
pip install xgboost contextilyRequirement already satisfied: xgboost in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (3.0.5)
Requirement already satisfied: contextily in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (1.6.2)
Requirement already satisfied: numpy in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from xgboost) (1.26.4)
Requirement already satisfied: nvidia-nccl-cu12 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from xgboost) (2.28.3)
Requirement already satisfied: scipy in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from xgboost) (1.16.1)
Requirement already satisfied: geopy in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from contextily) (2.4.1)
Requirement already satisfied: matplotlib in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from contextily) (3.10.0)
Requirement already satisfied: mercantile in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from contextily) (1.2.1)
Requirement already satisfied: pillow in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from contextily) (11.3.0)
Requirement already satisfied: rasterio in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from contextily) (1.4.3)
Requirement already satisfied: requests in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from contextily) (2.32.5)
Requirement already satisfied: joblib in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from contextily) (1.5.2)
Requirement already satisfied: xyzservices in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from contextily) (2025.4.0)
Requirement already satisfied: geographiclib<3,>=1.52 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from geopy->contextily) (2.1)
Requirement already satisfied: contourpy>=1.0.1 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from matplotlib->contextily) (1.3.3)
Requirement already satisfied: cycler>=0.10 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from matplotlib->contextily) (0.12.1)
Requirement already satisfied: fonttools>=4.22.0 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from matplotlib->contextily) (4.59.2)
Requirement already satisfied: kiwisolver>=1.3.1 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from matplotlib->contextily) (1.4.9)
Requirement already satisfied: packaging>=20.0 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from matplotlib->contextily) (25.0)
Requirement already satisfied: pyparsing>=2.3.1 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from matplotlib->contextily) (3.2.3)
Requirement already satisfied: python-dateutil>=2.7 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from matplotlib->contextily) (2.9.0.post0)
Requirement already satisfied: six>=1.5 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from python-dateutil>=2.7->matplotlib->contextily) (1.17.0)
Requirement already satisfied: click>=3.0 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from mercantile->contextily) (8.2.1)
Requirement already satisfied: affine in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from rasterio->contextily) (2.4.0)
Requirement already satisfied: attrs in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from rasterio->contextily) (25.3.0)
Requirement already satisfied: certifi in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from rasterio->contextily) (2025.8.3)
Requirement already satisfied: cligj>=0.5 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from rasterio->contextily) (0.7.2)
Requirement already satisfied: click-plugins in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from rasterio->contextily) (1.1.1.2)
Requirement already satisfied: charset_normalizer<4,>=2 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from requests->contextily) (3.4.3)
Requirement already satisfied: idna<4,>=2.5 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from requests->contextily) (3.10)
Requirement already satisfied: urllib3<3,>=1.21.1 in /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages (from requests->contextily) (2.5.0)
Note: you may need to restart the kernel to use updated packages.
Training Dataset Generation¶
In this exercise, we will focus on 4 main classes:
artificial surfaces
agricultural areas
forests and seminatual areas
water bodies
The names of these classes comes from Corine Land Cover Level 0 classification scheme: https://
Sample points selection¶
Generate 4 geoJSON files containing at least 10 points for each class using the website: https://
Follow this procedure for each land cover class:
Open the link: https://
geojson .io / #map = 12 /46 .48882 /11 .33004 Switch to satellite view selecting “Standard Satellite” as a base layer in the bottom left corner
Select the Draw Point tool and place a point in an area corresponding to the class
Repeat until you have at least 10 points
Save the geoJSON file with the top-left toolbar
Repeat for the other classes
Once that you have your 4 .geojson files, upload them to JupyterLab via drag & drop.
If you don’t want to do this small mapping exercise and skip directly to the next part, we prepared already the necessary files for the area of Bolzano, Italy.
Read the points in Python¶
Modify the file paths to match the files that you generated
urban_points_file = "./geojson/urban.geojson"
crops_points_file = "./geojson/crop.geojson"
forest_points_file = "./geojson/forest.geojson"
water_points_file = "./geojson/water.geojson"Use GeoPandas to read the vector data. We also assign the corresponding Corine Land Cover code.
import geopandas as gpd
import numpy as np
df_urban = gpd.read_file(urban_points_file).assign(**{"label": 100})
df_crops = gpd.read_file(crops_points_file).assign(**{"label": 200})
df_forest = gpd.read_file(forest_points_file).assign(**{"label": 300})
df_water = gpd.read_file(water_points_file).assign(**{"label": 500})ERROR 1: PROJ: proj_create_from_database: Open of /home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/share/proj failed
Inspect a GeoDataFrame object containing the sample points
df_urban.head(3)Visualize a set of points to check if they are placed correctly on the corresponding class:
import contextily as cx
df_wm = df_forest.to_crs(epsg=3857) # Points placed on forests
ax = df_wm.plot(figsize=(10, 10), alpha=0.5, edgecolor="k")
cx.add_basemap(ax)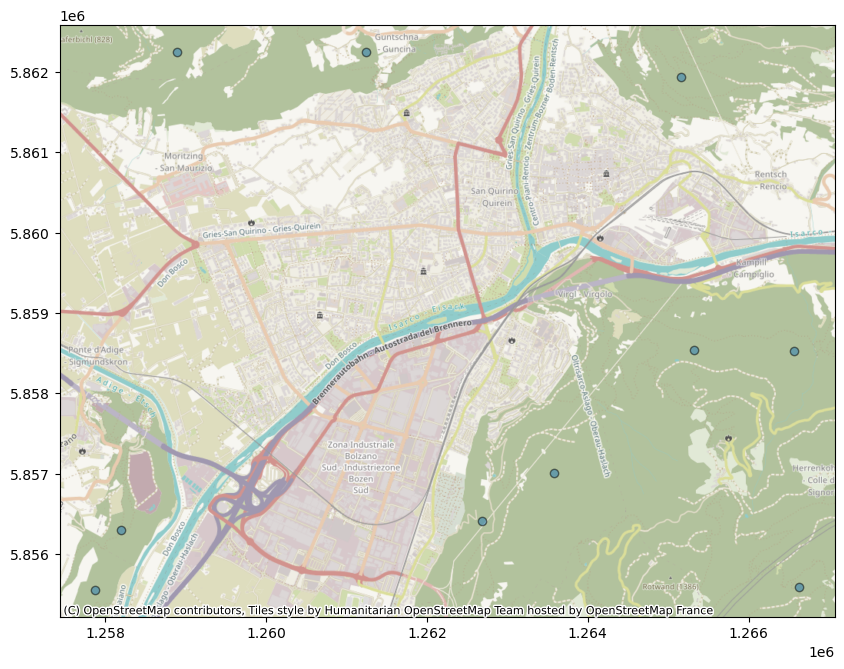
Satellite data retrieval¶
We will use optical Sentinel-2 data from the months of June, July and August 2022.
The data is available via the public EOPF Zarr STAC Catalog, available at https://
The xcube-eopf package will handle the STAC API requests and the datacube building parts.
from xcube.core.store import new_data_store
from xcube_resampling.utils import reproject_bboxstore = new_data_store("eopf-zarr")We would like to extract the bounds of the area containing our training points.
To do that we can combine the different GeoDataFrames into a single one and use the total_bounds method.
import pandas as pd
sample_points = pd.concat([df_urban, df_crops, df_forest, df_water])
bbox = sample_points.total_bounds
crs_utm = "EPSG:32632" # If you select the points in a different part of the World, you might need to use a different CRS
bbox_utm = reproject_bbox(bbox, "EPSG:4326", crs_utm)
bbox_utm(676214.7067171357, 5147669.36589368, 682626.7086097527, 5153320.106294904)ds = store.open_data(
data_id="sentinel-2-l2a",
bbox=bbox_utm,
time_range=["2025-06-01", "2025-08-31"],
spatial_res=10,
crs=crs_utm,
variables=[
"b04",
"b03",
"b02",
"b08",
# "scl"
], # b04 = red, b03 = green, b02 = blue, b08 = near infrared
)
dsPerform a temporal aggregation to get a monthly median of the data.
The median is capable to filter out data extremes, like clouds.
However, to get the best result is advisable to create and apply a cloud mask using the Scene Classification Layer, can you add this step before computing the median?
You can have a look at the example provided here: https://
Remember to add the “scl” band when creating the datacube and to remove it before performing the feature extraction.
# Apply the cloud mask to the data here:monthly_median = ds.groupby("time.month").median().to_dataarray(dim="bands")
monthly_medianThe default area above Bolzano corresponds to a manageable datacube which can fit easily in memory and therefore we will now load it in memory to speed up the next steps.
monthly_median = monthly_median.compute()/home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages/dask/_task_spec.py:758: RuntimeWarning: All-NaN slice encountered
return self.func(*new_argspec, **kwargs)
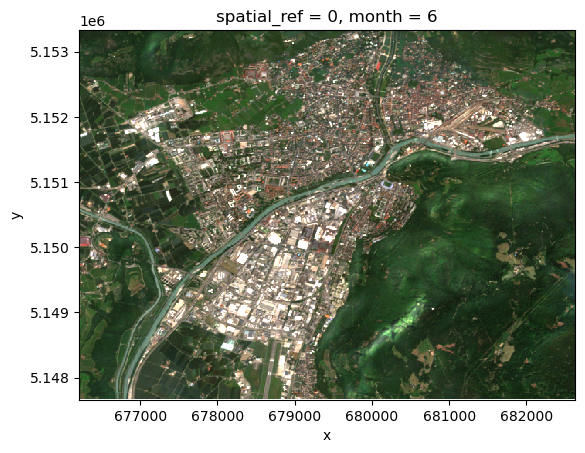
Visualize a sample image of the selected ones:
monthly_median.loc[dict(bands=["b04", "b03", "b02"])][:, 0].plot.imshow(
vmin=0, vmax=0.25
)
Predictors/Features extraction¶
Given the selected sample points and the Sentinel-2 data, we can now extract the data values corresponding to each sample point.
Each data array extracted will become a so-called predictor or feature vector.
Points - Data projection alignment¶
The geoJSON files contain points with coordinates expressed in lat/lon (WGS84 / EPSG:4326 coordinate system).
However, the data doesn’t necessarily has the same projection. Therefore, we reproject points to match data projection.
monthly_median.rio.crsCRS.from_wkt('PROJCS["WGS 84 / UTM zone 32N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563]],PRIMEM["Greenwich",0],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",9],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32632"]]')sample_points_reproj = sample_points.to_crs(monthly_median.rio.crs)Feature Vector extraction¶
Given the reprojected points we can get the corresponding values and create feature vectors, which will be used for training.
We store the feature vectores in the same GeoDataFrame as the sampled points:
predictors = []
for idx, s in enumerate(sample_points_reproj.iterrows()):
point = s[1]["geometry"]
data_values = monthly_median.sel(x=point.x, y=point.y, method="nearest").values
feature_vector = np.ndarray.flatten(data_values)
predictors.append(feature_vector)
sample_points_reproj = sample_points_reproj.assign(**{"features": predictors})
sample_points_reprojWe perform some operations to:
Get the feature vector in the desired shape: NxM with N as the number of points and M the number of features (in this case bands*time)
Get the corresponding label vector with the correct format. The ML library we are using is expecting labels with values starting from 0 and therefore we have to map them.
dict_mapping = {}
[
dict_mapping.update({x: lbl})
for x, lbl in enumerate(np.unique(sample_points_reproj["label"].values))
]
dict_mapping{0: 100, 1: 200, 2: 300, 3: 500}from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
y_train = le.fit_transform(sample_points_reproj["label"].values)
x_train = np.asarray([x for x in sample_points_reproj["features"].values])Random Forest¶
We use the xgboost ML library with its Random Forest implementation.
For more info please read the documentation here:
and for the colsample_by* hyperparameters explanation check this nice article here:
https://
Model training¶
from xgboost import XGBRFClassifier
model = XGBRFClassifier(n_estimators=100, subsample=0.9)
model.fit(x_train, y_train)Model prediction¶
To speed up the prediction process, we parallelize using xarray apply_ufunc, which allows to apply the same function call to multiple data chunks in parallel using Dask.
import xarray as xr
def predict_rf(x):
d = np.reshape(x, (x.shape[0] * x.shape[1], x.shape[2] * x.shape[3]))
pred = model.predict(d)
return np.reshape(pred, (x.shape[0], x.shape[1]))
def predict(obj, dims):
return xr.apply_ufunc(
predict_rf,
obj,
input_core_dims=[dims],
dask="parallelized",
output_dtypes=[int],
)
predicted = predict(monthly_median, ["bands", "month"])Prediction Evaluation¶
Now that our model has been trained and the prediction generated based on it, we can evaluate the result.
We didn’t do any split of the training set due to the limited number of samples, so we don’t have any test or validation set to use.
For this simple example, please evaluate the result using QGIS (or equivalent GIS software) and a satellite layer as a reference.
import matplotlib.pyplot as plt
import matplotlib.colors as mcolors
import matplotlib.patches as mpatches
import numpy as np
class_labels = {0: "Urban", 1: "Crops", 2: "Forest", 3: "Water"}
# Define a colormap with as many colors as classes
cmap = mcolors.ListedColormap(["red", "orange", "green", "blue"])
# Define normalization: boundaries between class values
bounds = [-0.5, 0.5, 1.5, 2.5, 3.5] # half-way between class values
norm = mcolors.BoundaryNorm(bounds, cmap.N)
# Show the image
plt.imshow(predicted.values, cmap=cmap, norm=norm)
# Build custom legend
patches = [
mpatches.Patch(color=cmap(i), label=label)
for i, label in enumerate(class_labels.values())
]
plt.legend(handles=patches, bbox_to_anchor=(1.05, 1), loc="upper left")
plt.show()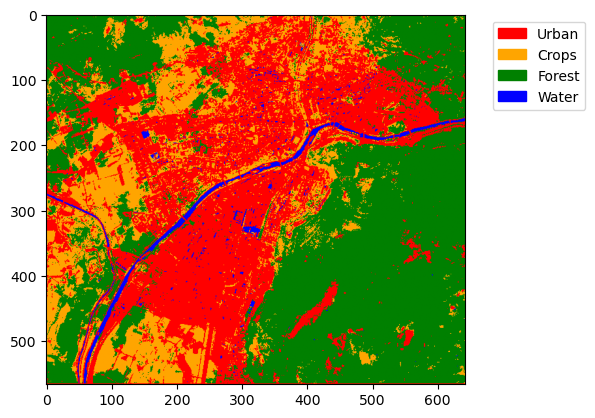
What’s next?¶
Now that you learned the basics, you can try to:
Create and apply a cloud mask to the data in the Satellite data retrieval section.
Apply the model to a new, unseen location and evaluate the results.
Apply the model to a different year compared to the training set.
