
Sentinel-1 L1 NRB product format prototype¶
Table of Contents¶
1. Introduction¶
In this notebook we will show an example of slc TOPSAR product and some easy usage examples.
Objectives¶
- Allow easy access to NRB product
- Provide ready-to-use datasets and data variable
- Allow users to open and manipulate data using both standard external tools and the EOPF
Relevant features¶
- Different polarizations, acquisition times are aligned and can be stacked together
- The coordinates associated to the data are the phisical coordinates
- The zarr product will adhere to CF convetion (this allow the user to open the prodoct with standards tools, such as xarray, exploiting properly the data coordinates).
- Phase and phase_model are optional variables. If present NRB product is equivalent to GSLC.
- STAC attributes (will be added in future)
Notes¶
This is a preliminary example of product
- Not all the metadata are included in these product prototype, but they will be included in the future.
- Among the excluded metadata, there are:
- qualityInformation
- The stac attrabutes are still to be defined.
- The name of the variables are preliminary and they may change in the future.
- Variable and group attributes will be refined in the future, including:
- long_name
- units
- standard names for coordinates
- The chunking is not defined yet.
- Products naming convention is to be defined.
- Reading from local and remote storage with EOPF is experimental (branch feat/coords_in_vars, commit 10a4f2e1) and not offically released.
import numpy as np
import xarray as xr
import datatree
import matplotlib.pyplot as plt3. Read products¶
Read local files with EOPF¶
The listed file do not work with the latest eopf library and therefore the next cells are commented out
# # local files
# !wget -q -r -nc -nH --cut-dirs=5 https://storage.sbg.cloud.ovh.net/v1/AUTH_8471d76cdd494d98a078f28b195dace4/sentinel-1-public/demo_product/nrb/ --no-parent -P /DIR_TO_WRITE_YOUR_PRODUCT/ --reject "index.html*"
# local_files = ['DIR_TO_WRITE_YOUR_PRODUCT/S01SIWNRB_20240106T170607_0028_A117_S000_5464A_DV.zarr',
# 'DIR_TO_WRITE_YOUR_PRODUCT/S01SIWNRB_20240205T051200_0027_A022_S000_5464A_DV.zarr',
# 'DIR_TO_WRITE_YOUR_PRODUCT/S01SIWNRB_20240205T051225_0028_A022_S000_5464A_DV.zarr']# ss = EOZarrStore(local_files[2])
# aa = ss.open()# eop = aa.load()Read remote files with xarray¶
# remote fles
files = [
"https://storage.sbg.cloud.ovh.net/v1/AUTH_8471d76cdd494d98a078f28b195dace4/sentinel-1-public/demo_product/nrb/S01SIWNRB_20240106T170607_0028_A117_S000_5464A_DV.zarr",
"https://storage.sbg.cloud.ovh.net/v1/AUTH_8471d76cdd494d98a078f28b195dace4/sentinel-1-public/demo_product/nrb/S01SIWNRB_20240205T051200_0027_A022_S000_5464A_DV.zarr",
"https://storage.sbg.cloud.ovh.net/v1/AUTH_8471d76cdd494d98a078f28b195dace4/sentinel-1-public/demo_product/nrb/S01SIWNRB_20240205T051225_0028_A022_S000_5464A_DV.zarr",
]dt = {}
for k, f in enumerate(files):
dt[k] = datatree.open_datatree(f, engine="zarr", chunks={})4. Product structure¶
EOPF¶
# eopXarray¶
dt[0]Loading...
measurements¶
dt[0].measurementsLoading...
measurements.nrb¶
dt[0].measurements.nrbLoading...
measurements.nlooks¶
dt[0].measurements.nlooksLoading...
conditions¶
conditions.refdem¶
dt[0].conditions.refdemLoading...
5. Examples of product usage¶
Selection with decimation factor 16 x 16¶
dt_sel = dt[0].measurements.isel(x=slice(None, None, 16), y=slice(None, None, 16))extent = [
dt_sel.x.min().values,
dt_sel.x.max().values,
dt_sel.y.min().values,
dt_sel.y.max().values,
]
fig, ((ax1, ax2, ax3), (ax4, ax5, ax6)) = plt.subplots(
nrows=2, ncols=3, figsize=(20, 20)
)
f = ax1.imshow(
dt_sel.nrb.sel(polarization="VV", time=np.datetime64("2024-01-06", "ns"))
* dt_sel.gamma_ref,
vmin=0,
vmax=2,
origin="lower",
extent=extent,
)
ax1.set_xlabel("east [m]", fontsize=10)
ax1.set_ylabel("north [m]", fontsize=10)
ax1.set_title("Sigma nought", fontsize=10)
ax1.grid()
fig.colorbar(f, ax=ax1, orientation="horizontal")
f = ax2.imshow(dt_sel.gamma_ref, vmin=0, vmax=2, origin="lower", extent=extent)
ax2.set_xlabel("east [m]", fontsize=10)
ax2.set_ylabel("north [m]", fontsize=10)
ax2.set_title("Gamma flattening", fontsize=10)
ax2.grid()
fig.colorbar(f, ax=ax2, orientation="horizontal")
f = ax3.imshow(dt_sel.inc, origin="lower", extent=extent)
ax3.set_xlabel("east [m]", fontsize=10)
ax3.set_ylabel("north [m]", fontsize=10)
ax3.set_title("Incidence angle", fontsize=10)
ax3.grid()
fig.colorbar(f, ax=ax3, orientation="horizontal")
f = ax4.imshow(
dt_sel.nrb.sel(polarization="VV", time=np.datetime64("2024-01-06", "ns")),
vmin=0,
vmax=2,
origin="lower",
extent=extent,
)
ax4.set_xlabel("east [m]", fontsize=10)
ax4.set_ylabel("north [m]", fontsize=10)
ax4.set_title("Gamma nought VV", fontsize=10)
ax4.grid()
fig.colorbar(f, ax=ax4, orientation="horizontal")
f = ax5.imshow(
dt_sel.nrb.sel(polarization="VH", time=np.datetime64("2024-01-06", "ns")),
vmin=0,
vmax=0.5,
origin="lower",
extent=extent,
)
ax5.set_xlabel("east [m]", fontsize=10)
ax5.set_ylabel("north [m]", fontsize=10)
ax5.set_title("Gamma nought VH", fontsize=10)
ax5.grid()
fig.colorbar(f, ax=ax5, orientation="horizontal")
im1 = ax6.imshow(
dt_sel.nrb.sel(polarization="VV", time=np.datetime64("2024-01-06", "ns")),
cmap=plt.cm.Reds,
interpolation="nearest",
vmin=0,
vmax=2,
origin="lower",
extent=extent,
)
im1 = ax6.imshow(
dt_sel.nrb.sel(polarization="VH", time=np.datetime64("2024-01-06", "ns")),
cmap=plt.cm.Greens,
interpolation="nearest",
vmin=0,
vmax=0.3,
alpha=0.8,
origin="lower",
extent=extent,
)
# f = ax6.imshow(wrap(dt_sel.phase.sel(polarization='VV', time=np.datetime64('2024-01-06', 'ns'))), vmin=-np.pi, vmax=np.pi, origin='lower', extent=extent)
ax6.set_xlabel("east [m]", fontsize=10)
ax6.set_ylabel("north [m]", fontsize=10)
ax6.set_title("Phase", fontsize=10)
ax6.grid()
fig.colorbar(im1, ax=ax6, orientation="horizontal")
fig.tight_layout()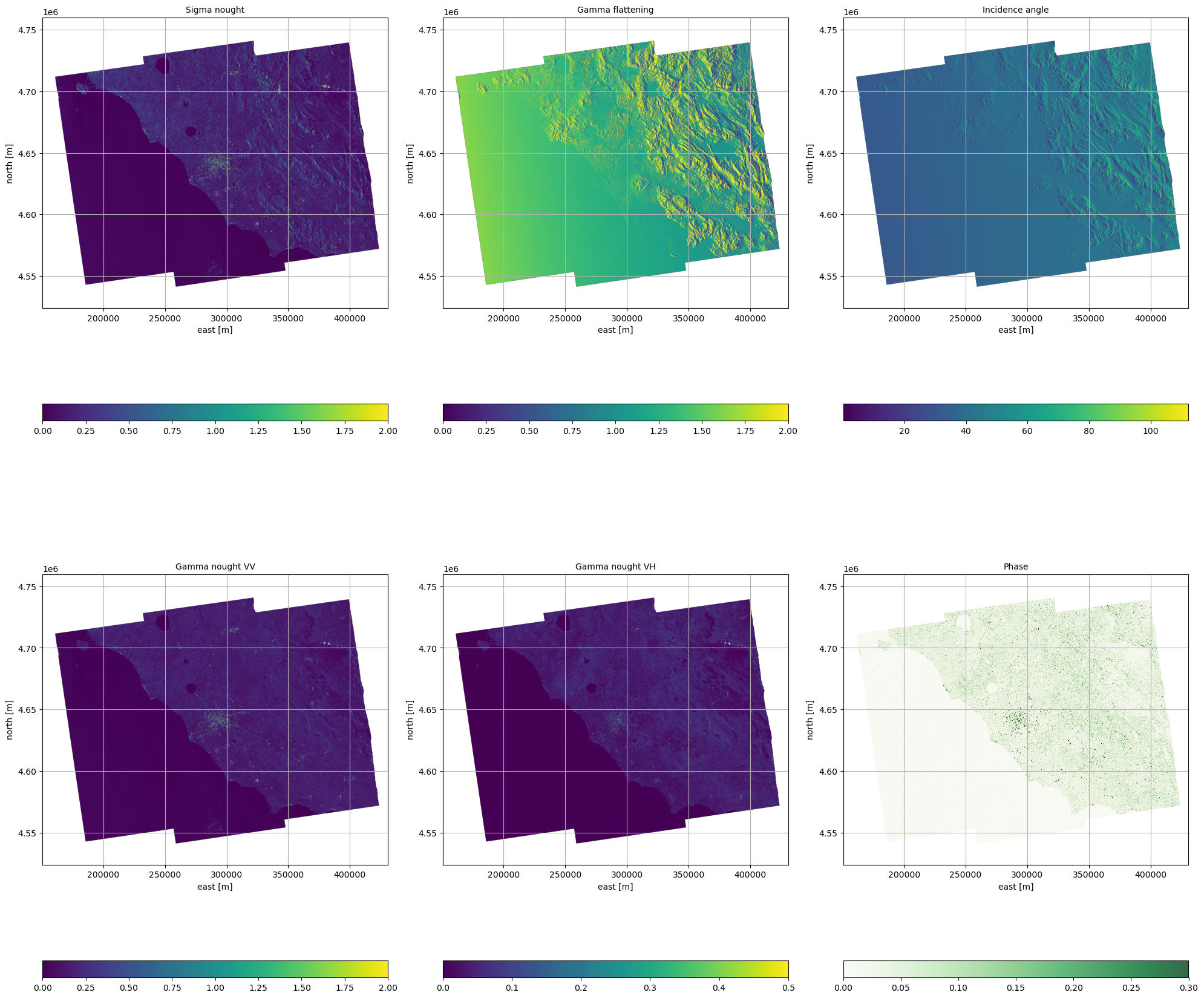
/home/mclaus@eurac.edu/micromamba/envs/eopf-zarr/lib/python3.11/site-packages/dask/core.py:127: RuntimeWarning: invalid value encountered in divide
return func(*(_execute_task(a, cache) for a in args))
Mix of different polarizations¶
# rescaling for visualization purpose
vv_max = (
dt_sel.nrb.sel(polarization="VV", time=np.datetime64("2024-01-06", "ns"))
.compute()
.quantile(0.9, skipna=True)
)
vh_max = (
dt_sel.nrb.sel(polarization="VH", time=np.datetime64("2024-01-06", "ns"))
.compute()
.quantile(0.9, skipna=True)
)
r = (
dt_sel.nrb.sel(polarization="VV", time=np.datetime64("2024-01-06", "ns")).drop(
["polarization", "time"]
)
/ vv_max
)
g = (
dt_sel.nrb.sel(polarization="VH", time=np.datetime64("2024-01-06", "ns")).drop(
["polarization", "time"]
)
/ vh_max
)
b = 2 * np.abs(r - g) / (r + g)/tmp/ipykernel_1126868/4188562811.py:13: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
dt_sel.nrb.sel(polarization="VV", time=np.datetime64("2024-01-06", "ns")).drop(
/tmp/ipykernel_1126868/4188562811.py:19: DeprecationWarning: dropping variables using `drop` is deprecated; use drop_vars.
dt_sel.nrb.sel(polarization="VH", time=np.datetime64("2024-01-06", "ns")).drop(
# concatenation of different bands
rgb = xr.concat([r, g, b], dim="rgb")
rgb = xr.where(rgb > 1, 1, rgb)
plt.figure(figsize=(12, 8))
rgb.plot.imshow(rgb="rgb")
plt.grid()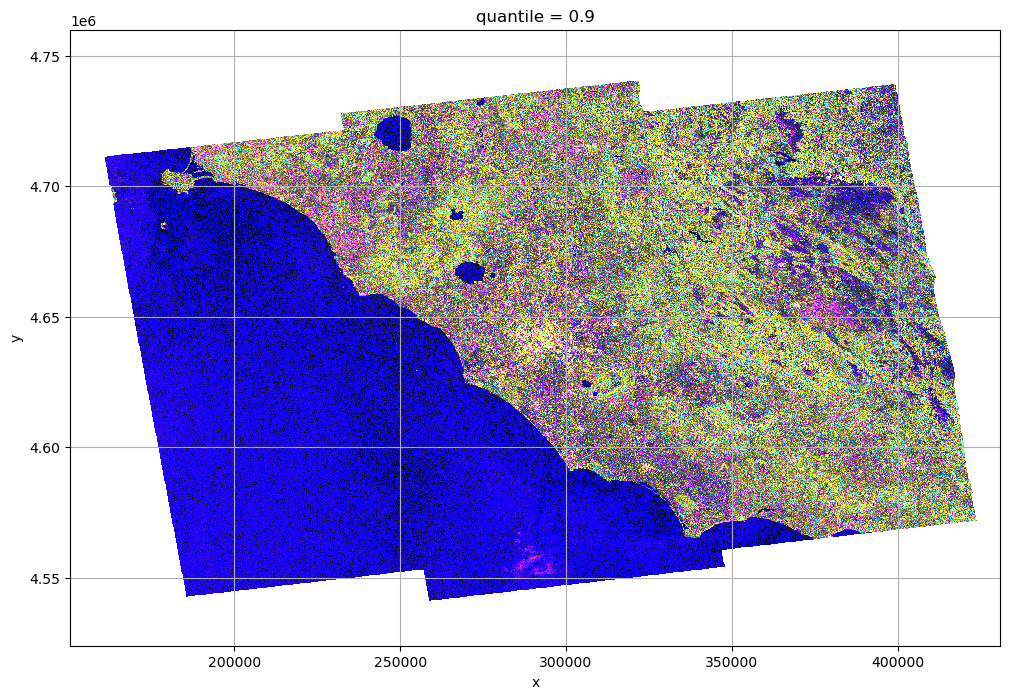
Selection based on geographic coordinates¶
dt_sel = []
for k, v in dt.items():
dt_sel.append(
v.measurements.sel(x=slice(260005, 360005), y=slice(4500025, 4800025))
.to_dataset()
.chunk(1024)
)dt_sel[0]Loading...
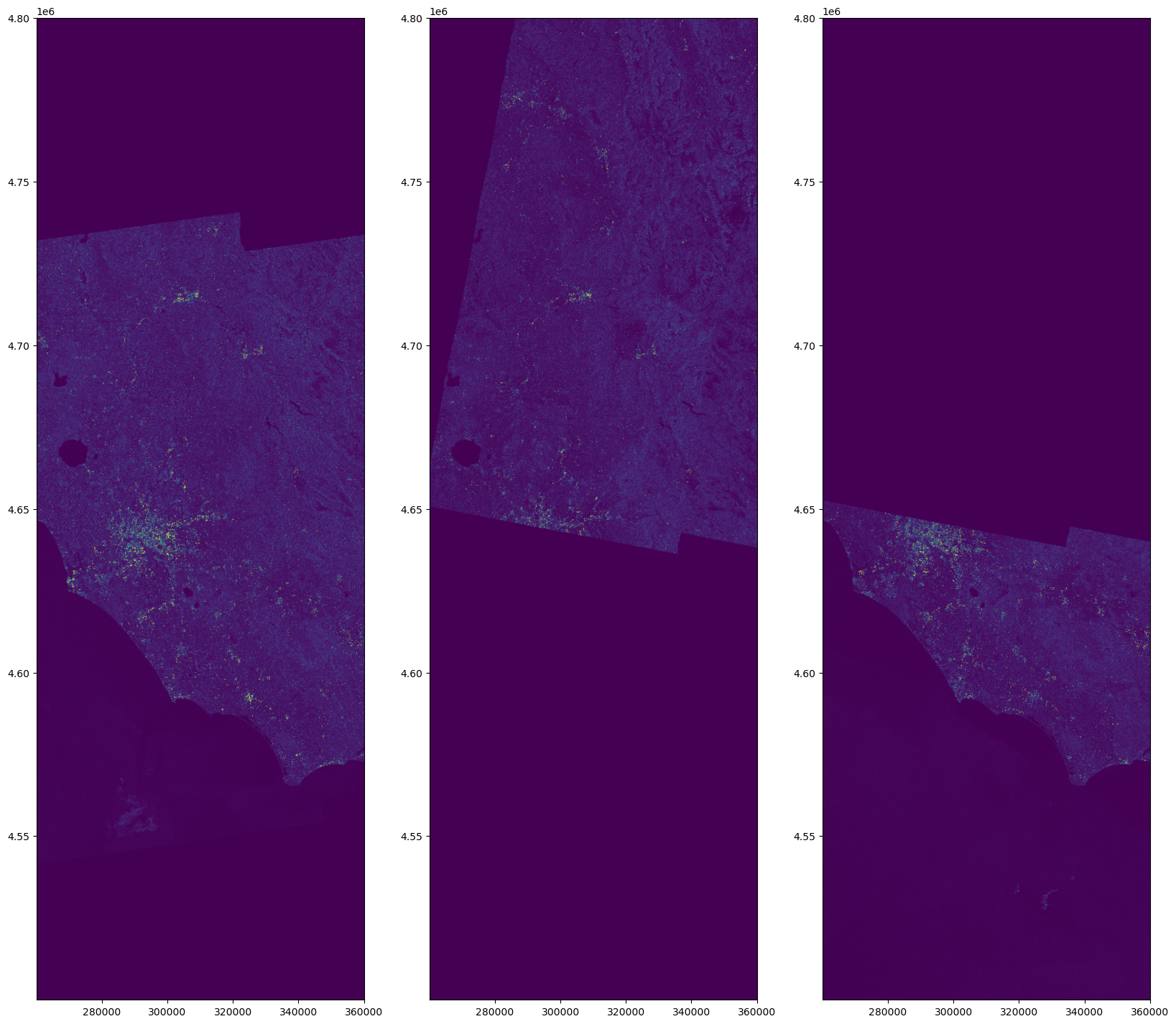
Mosaic¶
dt_align = xr.align(*dt_sel, join="outer", fill_value=0)nrb, w = [], []
for i in dt_align:
a = i.nrb.sel(polarization="VV").sum(dim="time")
a = a.where(np.isfinite(a), other=0)
nrb.append(a)
a = i.nlooks
a = a.where(np.isfinite(a), other=0)
w.append(a)nlooks = w[0] + w[1] + w[2]
mosaic = (nrb[0] * w[0] + nrb[1] * w[1] + nrb[2] * w[2]) / nlooksextent = [
mosaic.x.min().values,
mosaic.x.max().values,
mosaic.y.min().values,
mosaic.y.max().values,
]
fig, (ax1, ax2, ax3) = plt.subplots(nrows=1, ncols=3, figsize=(20, 20))
im1 = ax1.imshow(
nrb[0].isel(x=slice(None, None, 16), y=slice(None, None, 16)),
interpolation="bilinear",
vmin=0,
vmax=2,
origin="lower",
extent=extent,
)
im2 = ax2.imshow(
nrb[1].isel(x=slice(None, None, 16), y=slice(None, None, 16)),
interpolation="bilinear",
vmin=0,
vmax=2,
origin="lower",
extent=extent,
)
im3 = ax3.imshow(
nrb[2].isel(x=slice(None, None, 16), y=slice(None, None, 16)),
interpolation="bilinear",
vmin=0,
vmax=2,
origin="lower",
extent=extent,
)
extent = [
mosaic.x.min().values,
mosaic.x.max().values,
mosaic.y.min().values,
mosaic.y.max().values,
]
fig, ax1 = plt.subplots(nrows=1, ncols=1, figsize=(20, 20))
im1 = ax1.imshow(
mosaic.isel(x=slice(None, None, 2), y=slice(None, None, 2)),
cmap=plt.cm.gray,
interpolation="bilinear",
vmin=0,
vmax=1,
origin="lower",
extent=extent,
)
ax1.set_xlabel("east [m]", fontsize=10)
ax1.set_ylabel("north [m]", fontsize=10)
ax1.set_title("Gamma nought VV Mosaic", fontsize=10)
ax1.grid()
fig.colorbar(im1, ax=ax1, orientation="vertical")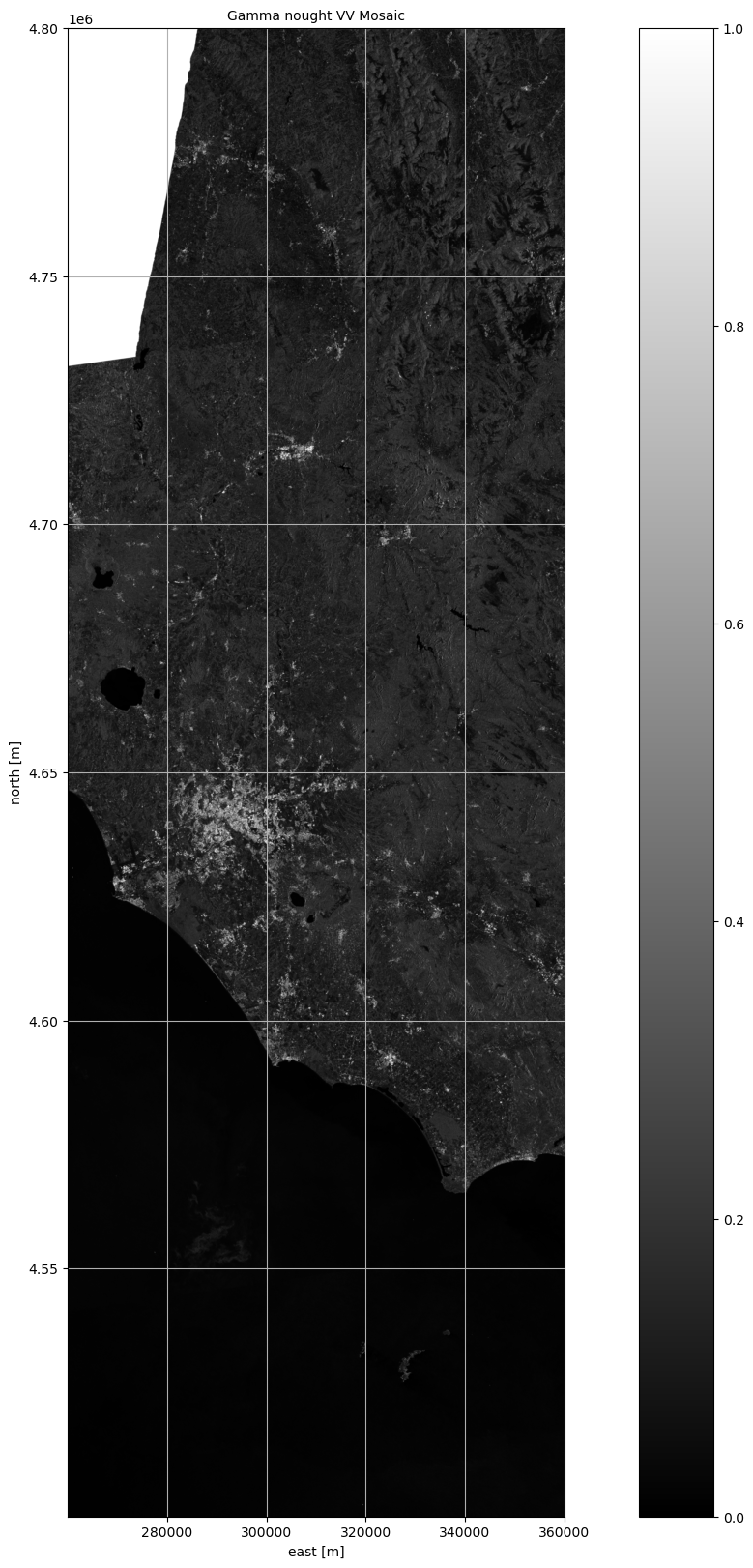
mosaicLoading...
6. Save & re-read derived products¶
Save & re-read mosaic with x-array from remote storage¶
Save & re-read mosaic with x-array from local storage¶
f = "DIR_TO_WRITE_YOUR_PRODUCT/S1A_NRB_VV_UTM33_260005_360005_4500025_4800025.zarr"
mosaic.chunk(1024).to_dataset(name="nrb").to_zarr(f, group="measurements", mode="w")
nlooks.chunk(1024).to_dataset(name="nlooks").to_zarr(f, group="quality", mode="a")
dt_mosaic = datatree.open_datatree(f, engine="zarr")
dt_mosaicRead mosaic with EOPF from local storage¶
# xs = EOZarrStore(
# "DIR_TO_WRITE_YOUR_PRODUCT/S1A_NRB_VV_UTM33_260005_360005_4500025_4800025.zarr"
# )
# ss = xs.open()# eop = ss.load()
# eop