STAC-Based Exploration and Visualization of Sentinel-2 and Sentinel-1 Data
Learn how to access Sentinel-2 and Sentinel-1 images via the EOPF STAC Catalog using pystac-client

Table of Contents¶
Run this notebook interactively with all dependencies pre-installed
Introduction¶
This notebook provides a comprehensive walkthrough for accessing, filtering, and visualizing Sentinel-1 and Sentinel-2 satellite imagery using a STAC (SpatioTemporal Asset Catalog) interface. Leveraging the pystac-client, xarray, and matplotlib libraries, the notebook demonstrates how to:
Connect to a public EOPF STAC catalog hosted at https://
stac .core .eopf .eodc .eu. Perform structured searches across Sentinel-2 L2A and Sentinel-1 GRD collections, filtering by spatial extent, date range, cloud cover, and orbit characteristics.
Retrieve and load data assets directly into xarray for interactive analysis.
Visualize Sentinel-2 RGB composites along with pixel-level cloud coverage masks.
Render Sentinel-1 backscatter data (e.g., VH polarization) for selected acquisitions.
Setup¶
Start importing the necessary libraries
import matplotlib.colors as mcolors
import matplotlib.pyplot as plt
import numpy as np
import pystac_client
import xarray as xr
from pystac_client import CollectionSearch
from matplotlib.gridspec import GridSpecSTAC Collection Discovery¶
Using CollectionSearch to list available STAC collections
# Initialize the collection search
search = CollectionSearch(
url="https://stac.core.eopf.eodc.eu/collections", # STAC /collections endpoint
)
# Retrieve all matching collections (as dictionaries)
for collection_dict in search.collections_as_dicts():
print(collection_dict["id"])sentinel-2-l2a
sentinel-3-olci-l2-lfr
sentinel-3-slstr-l2-lst
sentinel-1-l2-ocn
sentinel-1-l1-grd
sentinel-2-l1c
sentinel-1-l1-slc
sentinel-3-slstr-l1-rbt
sentinel-3-olci-l1-efr
sentinel-3-olci-l1-err
sentinel-3-olci-l2-lrr
Sentinel-2 Item Search¶
Querying the Sentinel-2 L2A collection by bounding box, date range, and cloud cover
catalog = pystac_client.Client.open("https://stac.core.eopf.eodc.eu")
# Search with cloud cover filter
items = list(
catalog.search(
collections=["sentinel-2-l2a"],
bbox=[7.2, 44.5, 7.4, 44.7],
datetime=["2025-01-30", "2025-05-01"],
query={"eo:cloud_cover": {"lt": 20}}, # Cloud cover less than 20%
).items()
)
print(items)[<Item id=S2B_MSIL2A_20250430T101559_N0511_R065_T32TLQ_20250430T131328>, <Item id=S2C_MSIL2A_20250425T102041_N0511_R065_T32TLQ_20250425T155812>, <Item id=S2C_MSIL2A_20250418T103041_N0511_R108_T32TLQ_20250418T160655>, <Item id=S2C_MSIL2A_20250405T102041_N0511_R065_T32TLQ_20250405T175414>]
Quicklook Visualization for Sentinel-2¶
We can use the RGB quicklook layer contained in the Sentinel-2 EOPF Zarr product for a visualization of the content:
item = items[0] # extracting the first item
ds = xr.open_dataset(
item.assets["product"].href,
**item.assets["product"].extra_fields["xarray:open_datatree_kwargs"],
) # The engine="eopf-zarr" is already embedded in the STAC metadata
ds.quality_l2a_quicklook_r60m_tci.plot.imshow(rgb="quality_l2a_quicklook_r60m_band")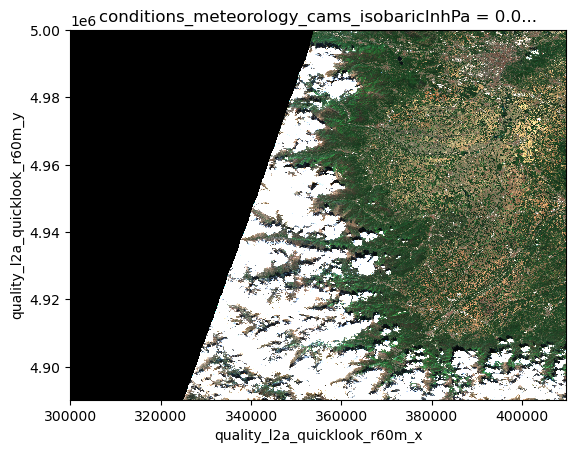
We can view side by side the RGB quicklook and the SCL (Scene Classification Layer) for all the returned STAC Items:
def visualize_items_table(items):
"""
Visualize multiple Sentinel-2 items with scene IDs centered above each row,
followed by two columns (RGB and SCL).
Parameters:
items (list): List of STAC items
"""
num_items = len(items)
row_height = 7 # Approximate height per item
fig_height = min(row_height * num_items, 24)
fig = plt.figure(figsize=(14, fig_height))
# Create height ratios: 0.3 for title row, 1.7 for image row (per item)
height_ratios = []
for _ in range(num_items):
height_ratios.extend([0.3, 1.7]) # first is title, second is images
gs = GridSpec(
num_items * 2, 2, height_ratios=height_ratios, hspace=0.1, wspace=0.05
)
# Create colorbar axis (positioned below all plots)
scl_cbar_ax = fig.add_axes([0.3, 0.03, 0.4, 0.015]) # [left, bottom, width, height]
for i, item in enumerate(items):
try:
# Add centered title for the row
title_ax = fig.add_subplot(gs[i * 2, :]) # Span both columns
title_text = f"{item.id} (Cloud: {item.properties['eo:cloud_cover']:.1f}%)"
title_ax.text(
0.5,
0.5,
title_text,
ha="center",
va="center",
fontsize=10,
fontweight="bold",
)
title_ax.axis("off")
# Load the dataset
ds = xr.open_dataset(
item.assets["product"].href,
**item.assets["product"].extra_fields["xarray:open_datatree_kwargs"],
)
# RGB Column
ax1 = fig.add_subplot(gs[i * 2 + 1, 0])
try:
if "quality_l2a_quicklook_r60m_tci" in ds:
ds.quality_l2a_quicklook_r60m_tci.plot.imshow(ax=ax1)
ax1.set_title("True Color (TCI)", pad=5, fontsize=9)
else:
ax1.text(
0.5,
0.5,
"No RGB quicklook",
ha="center",
va="center",
fontsize=8,
)
except Exception:
ax1.text(0.5, 0.5, "RGB Error", ha="center", va="center", fontsize=8)
ax1.axis("off")
# SCL Column
ax2 = fig.add_subplot(gs[i * 2 + 1, 1])
try:
if "conditions_mask_l2a_classification_r60m_scl" in ds:
scl = ds.conditions_mask_l2a_classification_r60m_scl.compute()
# Apply classification attributes
scl.attrs.update(
{
"flag_values": [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11],
"flag_meanings": (
"no_data sat_or_defect_pixel topo_casted_shadows cloud_shadows vegetation not_vegetation water unclassified cloud_medium_prob cloud_high_prob thin_cirrus snow_or_ice"
),
"flag_colors": (
"#000000 #ff0000 #2f2f2f #643200 #00a000 #ffe65a #0000ff #808080 #c0c0c0 #ffffff #64c8ff #ff96ff"
),
}
)
cmap = mcolors.ListedColormap(scl.attrs["flag_colors"].split(" "))
norm = mcolors.BoundaryNorm(
boundaries=np.arange(13) - 0.5, ncolors=12 # For 12 classes
)
img_scl = scl.plot.imshow(
ax=ax2, cmap=cmap, norm=norm, add_colorbar=False
)
ax2.set_title("Scene Classification", pad=5, fontsize=9)
# Create colorbar once
if i == 0:
cbar = fig.colorbar(
img_scl,
cax=scl_cbar_ax,
orientation="horizontal",
ticks=np.arange(12),
)
class_labels = [
label.replace("_", " ").title()
for label in scl.attrs["flag_meanings"].split()
]
cbar.ax.set_xticklabels(
class_labels, rotation=45, ha="right", fontsize=8
)
cbar.set_label("Land Cover Classes", fontsize=9)
else:
ax2.text(
0.5, 0.5, "No SCL layer", ha="center", va="center", fontsize=8
)
except Exception:
ax2.text(0.5, 0.5, "SCL Error", ha="center", va="center", fontsize=8)
ax2.axis("off")
except Exception as e:
print(f"Error processing item {item.id}: {str(e)}")
continue
plt.tight_layout()
plt.subplots_adjust(bottom=0.07, top=0.95) # Adjust for colorbar and titles
plt.show()
visualize_items_table(items)/tmp/ipykernel_2583155/2474849155.py:126: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
plt.tight_layout()

Sentinel-1 GRD Item Search¶
Querying Sentinel-1 L1 GRD collection given bounding box, orbit direction and orbit track number.
# Load the STAC catalog
catalog = pystac_client.Client.open("https://stac.core.eopf.eodc.eu")
# Search for Sentinel-1 GRD items matching the metadata characteristics
items = list(
catalog.search(
collections=["sentinel-1-l1-grd"],
bbox=[29.922651, -9.648995, 32.569355, -7.391839],
datetime=[
"2025-09-09T00:00:00Z",
"2025-09-13T23:59:00Z",
],
query={
"sat:orbit_state": {"eq": "ascending"},
"sat:relative_orbit": {"eq": 174},
},
).items()
)
items[<Item id=S1A_IW_GRDH_1SDV_20250913T161934_20250913T161959_060971_079880_E055>,
<Item id=S1A_IW_GRDH_1SDV_20250913T161905_20250913T161934_060971_079880_AB93>]Sentinel-1 GRD Visualization¶
Opening the first STAC Item and plot the VH polarization
item = items[0]
ds = xr.open_dataset(
item.assets["product"].href,
**item.assets["product"].extra_fields["xarray:open_datatree_kwargs"],
variables="VH_measurements*",
)
plt.imshow(ds["VH_measurements_grd"][::10, ::10].to_numpy(), vmin=0, vmax=200)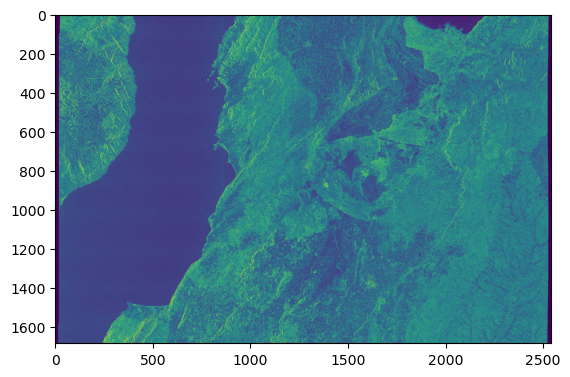
The Sentinel-1 visualization is off due to the EOPF CPM version used for the data conversion. The data must be converted with eopf >= 2.6.1.
print(item.assets["product"].href.split("/cpm_")[1].split("/")[0])v256